Welcome to the Baker Heart and Diabetes Institute Metabolomics Laboratory Portal. Here you will find links to our online resources, such as our genetic PheWebs and comprehensive overview of our lipidomics platform.
Comprehensive genetic analysis of the human lipidome. Using our advanced lipidomic methodology, we measured 596 lipid species measured in 4,492 individuals of the Busselton Health Study. See associations with over 13 million genetic variants.
Visit standard phewebCirculating lipoproteins are made up of thousands of lipid species and proteins. We have assessed genetic associations with lipid species, independent of clinical lipid traits (total cholesterol, HDL cholesterol and triglycerides).
Visit clinical lipid adjusted phewebA combined analysis of three independent cohorts, totalling more than 6,000 individuals. Incororating the Busselton Health Study, the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing, and the Alzheimer's Disease Neuroimaging Initiative.
Visit meta-analysis pheweb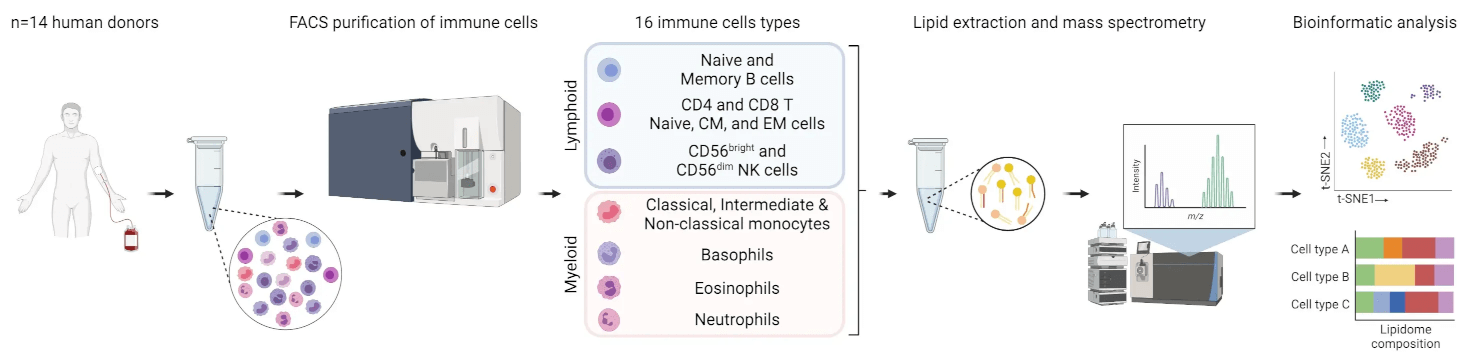
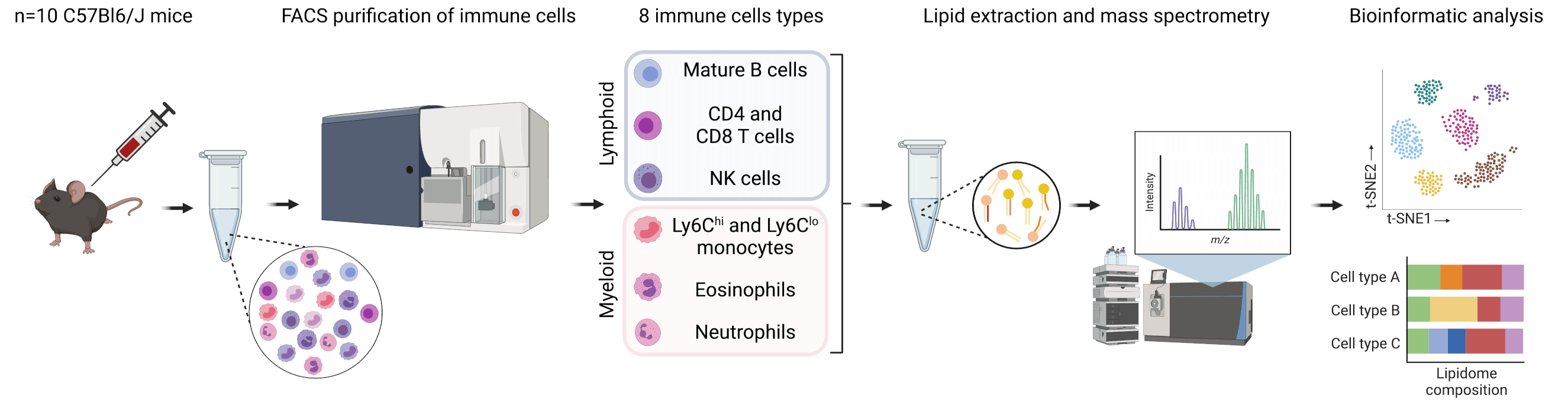
The Cellular Lipid Atlas is a comprehensive resource that maps the lipidome of cells within the human and mouse immune systems. The Cellular Lipid Atlas highlights that the lipid landscape is a defining feature of immune cell identity and physiology, providing valuable insights into immune cell function and health.
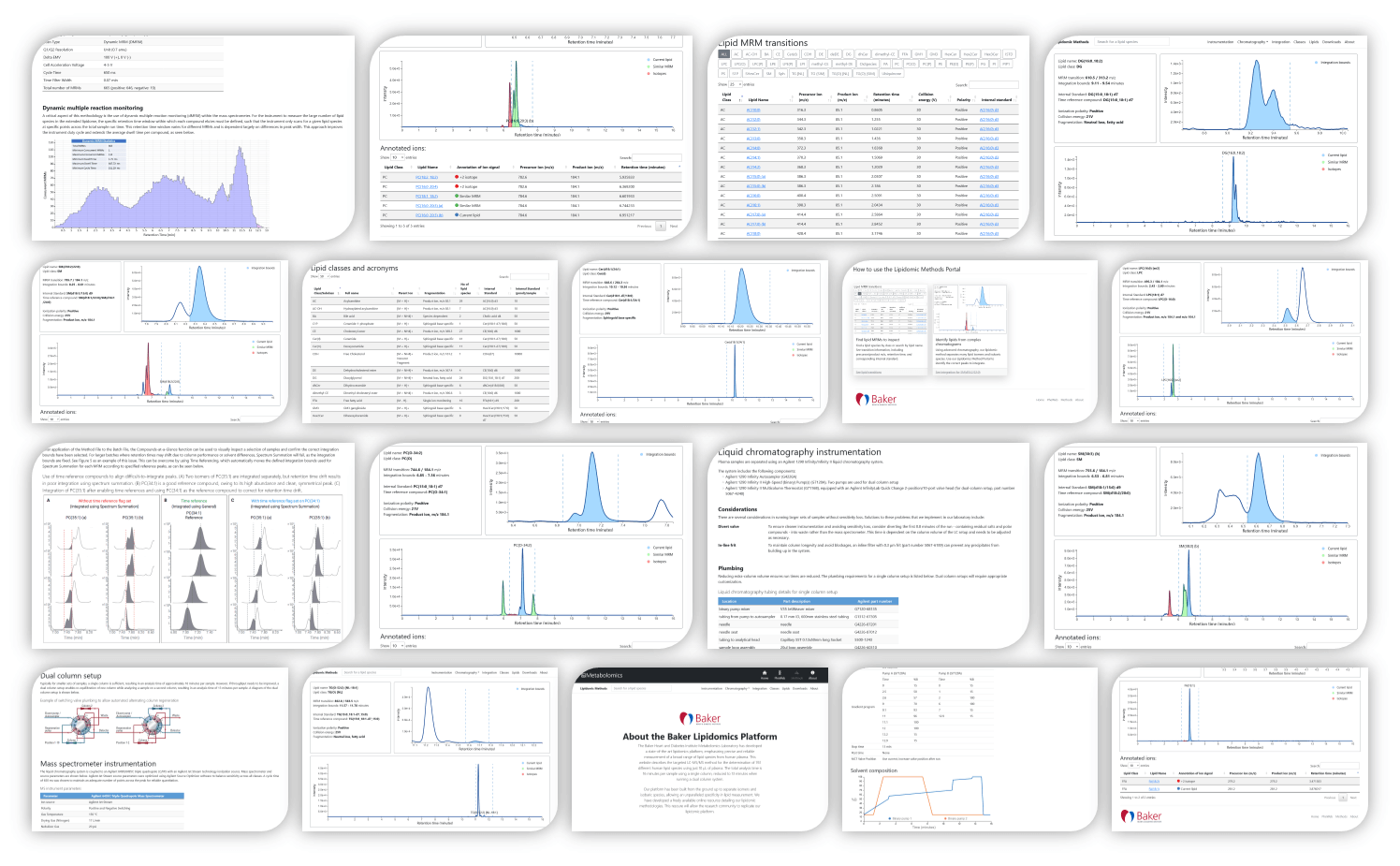
The Baker Heart and Diabetes Institute Metabolomics Laboratory has developed a state-of-the-art lipidomics platform, emphasizing precise and reliable measurement of a broad range of lipid species from human plasma. We have developed a freely available online resource detailing our lipidomic methodology. This resoure will allow the research community to replicate our lipidomic platform.